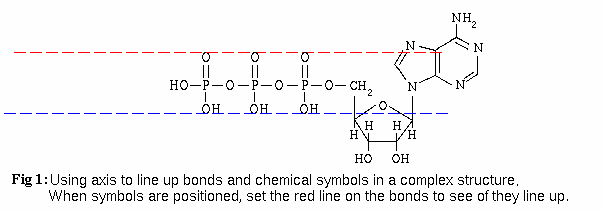
We start with a ring system and create 2 colored, dashed lines and a bunch of required symbols. We then use the dashed lines to align the single bonds and symbols like so:

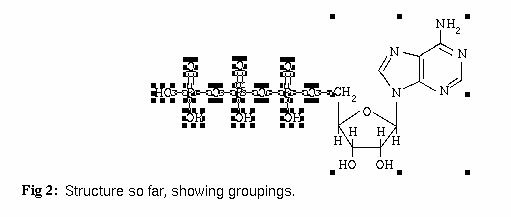
The last step, after selecting the completed structure, is to select Structure::Ungroup from the IVtools menu 2 or 3 times. You then reselect the completed structure and select Structure::Group from the IVtools menu. This will give you the result in Fig. 3 below.
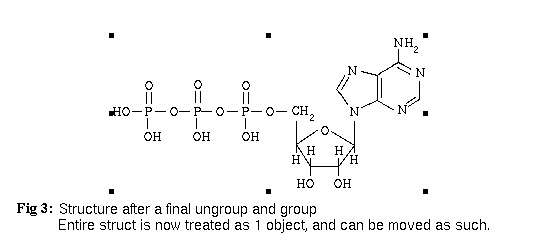
To include this structure in a technical paper or journal article, simply export it as encapsulated PostScript (EPS) by choosing File::Export Graphic. Click on "EPS", then click on "Export". Then insert the following before the \begin{document} line in your LaTeX document:
\input psfig.sty
To display the graphic, insert the following in the body of your LaTeX code:
\begin{figure}[htbp]
\psfig{file=file.ps,width=6in,height=3in} % see Kenneth Shultis, pp. 100-103
\end{figure}
Where file.ps is the name of your graphic; there cannot be any extraneous spaces inside the braces on the \psfig line. See Ken Shultis' book "LaTeX NOTES: Practical Tips for Preparing Technical Documents", ISBN: 0-13-120973-6
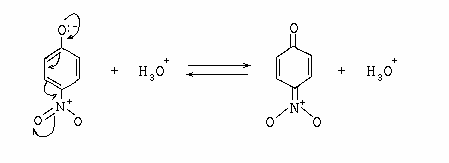
IVtools does Curvey-arrows
"Curvey arrow notation" is an official notation used by chemists for specifiying
chemical mechanisms by showing where the electrons go based on Lewis Acid-Base and
VSEPR (Valence Shell Electron Pair Repulsion) theories, and the Octet Rule.
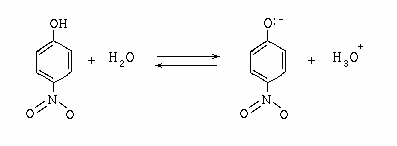
A very simple example of this is the resonance structure of 4-nitrophenol:
IVtools and heterocycles: The cyclical structures cyclobutane, cyclopentane, cyclohexane, benzene, cycloheptane, and cyclooctane were designed with removable angle vertexes making construction of heterocycles a snap:
The IVtools custom toolbar allows one to load custom toolbar configurations for different typesetting projects. Instead of loading all of the possible chemical widgets, and believe me there are alot, only load the ones required for the project at hand, or the ones you find most useful. To build a custom toolbar, write a toolbar script similar to this:
addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgets/c5-ring.idraw" :popen) addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgets/c6-ring.idraw" :popen) addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgets/phenyl.idraw" :popen) addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgets/c7-ring.idraw" :popen) addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgetsc/8-ring.idraw" :popen) addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgets/c6-boat.idraw" :popen) addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgets/c6-chair.idraw" :popen) addtool("/usr/guest/todd/graphics/IVTOOLS/ToolBars/Chem-Toolbar/widgets/t-butyl.idraw" :popen)
To load the desired toolbar click on Tools::Custom Tools. This will give a stripped down toolbar into which custom widgets can be inserted. To load the custom widgets, click into the Comdraw command window, and enter the following command:
run ("$PATH/file")
Where $PATH is the complete path to the custom toolbar script and "file" is the name of the script.
The chemistry widgets must be in idraw format to be added to the toolbar in this manner. To convert a widget to idraw format for use in the toolbar:
Below is a shot of the comdraw GUI with chem widgets loaded:
As an aside, I have also built a protein widget set, for use in amino-acid sequencing, building biomolecules etc. Here is a comparison of the first 15 amino-acid residues that comprise 3 different kinds of haemoglobin found in the sea lamprey, Petromyzon marinus:

[In the above example, the "spacing-widget.ps" template was used to line up the 3 amino-acid sequences]